Reproducible R
These notes are designed to accompany a presentation and workshop given by Alain Danet as an introduction to the use of the targets package for optimising large statistical computational pipelines in R. The corresponding slides are available here.
A problem in analysis is that we have bigger and bigger datasets – this means that if we`re running complicated models and creating plots, it can take a lot of computational time to run. Even analyses on small datasets can take up a lot of time if the models are particularly complex.
One of the key things that increases computational time is repetition. Rerunning an entire script every time you change a line, for example, can mean analyses take much longer than they need to. We often have the perception that it will be quick to rerun stuff, but in reality this often isn’t true!
It’s therefore convenient to have a tool that reduces computational time. The
targetspackage is one solution.A computational pipeline has a number of interconnected steps – changing something in one step can mean that some, but not necessarily all, other steps also need to be updated and rerun. If you have a really complicated pipeline, though, working out manually what other steps have become outdated can be a daunting task, so you might just end up rerunning everything. This is unnecessary and can drastically increase computational time.
targetsoffers a way to build a pipeline tool that allows you to keep track of which steps depend on other steps. If you update one step, targets automatically works out and tells you which other steps are now out of date and need to be rerun, Crucially, this allows you to rerun only the steps that you need, reducing computational time.The goal of targets is to move away from numbered imperative scripts (which can be misleading, because numbered scripts can`t represent complicated interdependencies) and instead embrace a workflow-based model.
The typical targets project structure consists of a master script, a folder containing all functions (e.g. loading data, filtering data, modelling, making plots etc.), and a folder containing all data.
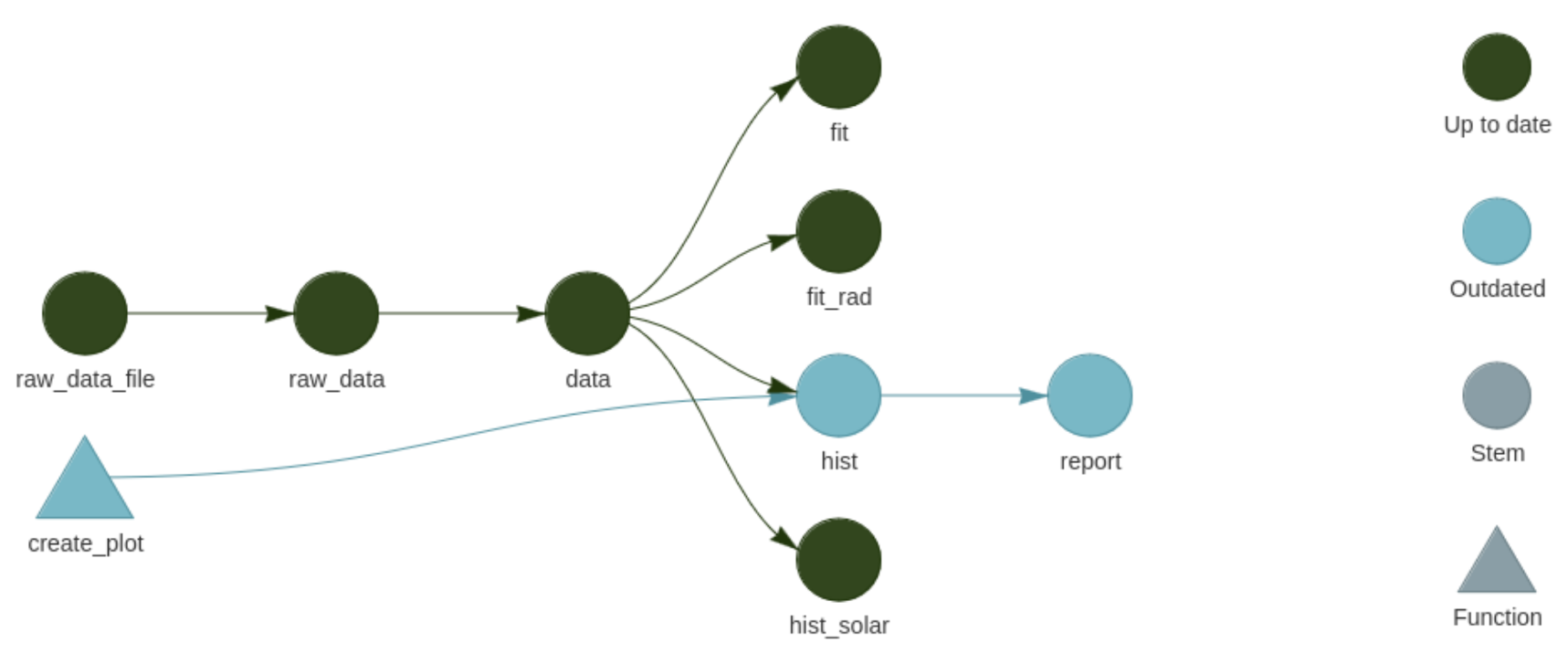 Example of a target pipeline where the modification of one function (
Example of a target pipeline where the modification of one function (create_plot() on the left) has outdated two other steps. Targets will rerun only two steps (hist and report) instead of the whole analysis.
Demonstration of minimal example by Will Landau
Use these notes and the slides to run through the example in Rstudio. First make sure you have installed the tidyverse, targets, tarchetypes, and biglm packages. Open the targets-minimal.Rproj file.
The _targets.R script
- A targets workflow consists of a list of
targets– think of these as the steps in your workflow. - The
_targets.Rfile is the master script which contains all the other steps. Open this script. The first step is to load up thetargetsandtarchetypespackages, which are needed to define the pipeline. - Note that targets runs the pipeline in a separate R session, so you need to tell it which packages to use with
tar_option_set()(just as you would load packages usinglibrary()in a normal R session). - The pipeline defines each step (loading in data, filtering, plotting etc) using the
tar_target()function. This creates a target object for each step. These are stored in a folder of target objects and are automatically saved. A pipeline is essentially a list of (skippable) target objects.- First argument in
tar_target()is the name of the object to be created. - Second argument is the code to create the object (e.g.
read_csv(raw_data_file, col_types = cols())to read in a .csv data file).
- First argument in
- Once it has a list of targets (from the master script), targets can understand the code and data dependencies.
The make.R script:
- Once we’ve got some steps in our targets pipeline, we can visualise it, check what needs to be updated, and update it. This needs to be done in a separate script (
make.R). - To view a table displaying all the target objects (with their names and associated commands), use the
tar_manifest()function. tar_visnetwork()visualises the objects, any functions needed to create them, and the interdependencies between them. It will tell you if each object is outdated and needs to be updated. You can click on each object to easily see its dependencies.- Outdated objects can be updated (
built) using thetar\_make()function tar_read()is used to read in data (without loading it in your current session) – this can be useful if you have, for example, a very big object that you want to do stuff with without loading it into your current session, or for viewing a plot. To actually load an object into your current session, use thetar_load()function.
Note that pipelines should be built up gradually – add one or two target objects at a time, so you can easily debug. Each time you add a new object, targets can check how this affects other objects in the workflow. tar_outdated() can be used to tell you which objects are now outdated (including the new objects). This can also be done by visualising.
- Running
tar\_make()` again will update only the outdated objects – visualising again (or running tar_outdated()) allows us to see that these objects have been updated. - The key point here is that tar_make() runs only the outdated objects, and skips any objects that don`t need to be updated, saving computational time
What if we need to change a function?
- In the
Rfolder in the example, there is a script calledfunctions.R - In this script, there is a function to create a plot – let`s change something in it (e.g. number of bins), save the script, and visualise the network again
- We can see that target recognises that the function has been changed, and both the function and every object that uses the function are shown as outdated
- This means you don`t have to read through your script to find every time the function has been used! And what if the data changes?
- Let’s go back to the
_targets.Rscript and change our data filtering step totar\_target(data, na.omit(raw_data))`, then save the script - Then when we visualise it, we can see that all the steps depending on the filtered data are now outdated. Again, we can update them with
tar_make()`.
R Markdown and targets
One of the good things about targets is that you can do it all via R Markdown. This means you can use it to produce an accessible, easily workflow including explanations of each step, the code used to perform it, and the corresponding outputs. See the excellent targets package user manual for a great example of the type of document you can produce using R Markdown.
In summary
Investing a little time into organising your work into a targets pipeline at the start of a project will save you a lot of time in the long run. Happy coding!
Alain Danet SESSION
coding tutorial reproducibility R
